Benzoxaborole as a new chemotype for carbonic anhydrase inhibition.
Alterio, V., Cadoni, R., Esposito, D., Vullo, D., Fiore, A.D., Monti, S.M., Caporale, A., Ruvo, M., Sechi, M., Dumy, P., Supuran, C.T., Simone, G., Winum, J.Y.(2016) Chem Commun (Camb) 52: 11983-11986
- PubMed: 27722534
- DOI: https://doi.org/10.1039/c6cc06399c
- Primary Citation of Related Structures:
5JQ0, 5JQT, 5LMD - PubMed Abstract:
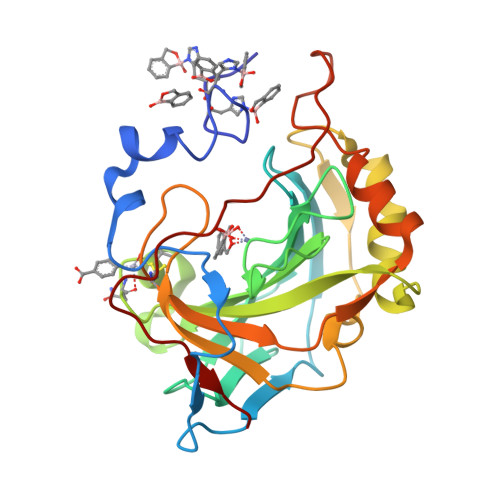
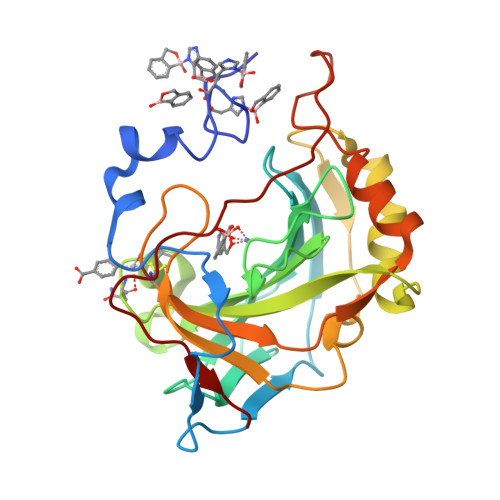
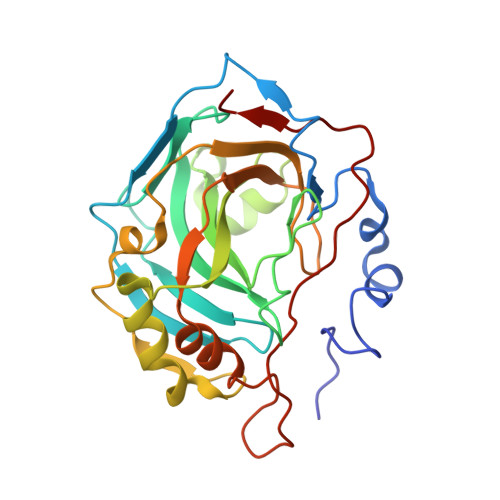
In this paper we report the synthesis of a series of benzoxaborole derivatives, their inhibition properties against some carbonic anhydrases (CAs), recognized as important drug targets, and the characterization of the binding mode of these molecules to the CA active site. Our data provide the first experimental evidence that benzoxaboroles can be efficiently used as CA inhibitors.
Organizational Affiliation:
Istituto di Biostrutture e Bioimagini-CNR, Naples, Italy. gdesimon@unina.it.